点击化学的TOP9应用
Click Chemistry由K.Barry Sharpless、Hartmuth C.Kolb和M.G.Finn于2001年提出,用于描述快速选择性反应或以可预测的方式相互“点击”以形成具有杂原子链(C-X-C)的生理稳定产物的反应。Click chemistry广泛用于生物分子、表面、颗粒和有机化合物的改性,具有许多优点1:
- 应用范围广泛;
- 模块化性质;
- 在“小量”和“大量”反应中均适用;
- 反应条件温和;
- 产品分离简单(几乎不需要纯化);
- 产率高,速度快;
- 无害副产品生成(遵循绿色化学的12项原则);
- 兼容性良好,尤其在生命系统中(允许生物分子的化学选择性修饰,几乎不受干扰)2。
在大约10种不同类型的点击反应中,有几种是在各种生命科学应用中使用最频繁,从“简单”的生物分子标记和检测到先进的CRISPER应用。有关更多信息,请参阅常见点击反应(Click Reactions)及其试剂系列。这里,我们重点介绍最重要的9种(最新)应用:
- 生物分子标记与检测
- (固&液相)生物分子修饰/连接
- 构建用于构效关系分析的类似物库
- 药物先导化合物发现
- 药物输送
- 材料优化(聚合物改性)
- 病毒研究探针
- CRISPER sgRNA合成和靶基因标记
- 新应用,包括“点击发布”
1.生物分子标记与检测
Click chemistry最有用的功能之一是它能够标记和可视化生物分子,如脂质3、肽4、聚糖5、糖蛋白6、核酸和合成分子7,8(如紫杉醇9),并且具有最小的生理干扰性(体外和体内)10。在进行标记的两步反应中,首先用双正交点击手柄(如炔烃或叠氮化物)标记目标生物分子(酶、代谢11,12或合成(请见图1)9,13)。然后当一个分子上有荧光或亲和基团的互补点击手柄与目标分子发生点击反应时,就会发生检测/可视化。
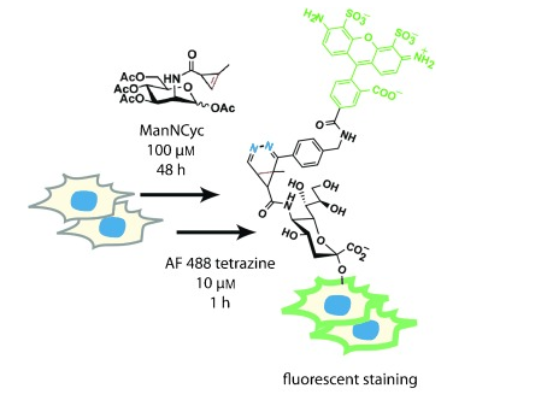
例如,在活体发育的斑马鱼中,表面聚糖以亚细胞分辨率被观察到;依靠基因编码的传统分子成像方法通常无法看见14。在这项研究中,Bertozzi等人将代谢糖工程与多色检测策略相结合,以揭示细胞表面表达、细胞内运输和整个斑马鱼胚胎发生过程中聚糖组织分布的差异。类似的研究也在小鼠中进行,以跟踪移植细胞和测定细胞对肽的摄取情况,这有助于结构-活性-通透性关系优化研究15。两个位点标记生物分子(称为双位点标记)有效的促进了复杂生物系统的研究16,17,18,19,20。
2.(固相和液相)生物分子修饰/连接
肽、核苷酸、小分子、超分子等都可以通过固相或液相点击化学进行修饰,几乎无需使用保护基团,也无需产品纯化3,21,22。总体来讲,固相合成更快,且需要更少的后处理,但是每种方法都各有优缺点23,24。
3. 类似物库的建设
类似物库可以通过点击化学快速可靠地构建,无需太多的合成工作,然后通过原位高通量筛选(HTS)来促进分子结构-活性关系(SAR)分析,这是优化和发现生物活性分子所必需的。已经有许多基于click(三唑)骨架的片段库(聚焦组合)通过此方法被构建出来25,例如Janus激酶抑制剂ruxolitinib衍生的三唑文库,它被用来评估JAK3抑制剂24。
4. 用于先导化合物发现的原位点击化学
原位点击化学是一种(动力学)靶点导向合成方法,Sharpless及其同事于2002年首次提出并应用于发现一种有效的乙酰胆碱酯酶抑制剂26。这种方法使用目标生物分子本身作为支架,如果使其足够接近并以适当的方向反应,则结合配体在其上进行咔哒反应。通过这种方式,可以从带有互补反应性官能团的片段库中筛选出能够与目标物形成稳定络合物的最佳配体27。无需事先对文库成员进行合成、纯化和生化评估,即可快速且经济高效地筛选大量化合物28,29。
碳酸酐酶30、HIV蛋白酶31、几丁酶32、核苷酸配体33、蛋白质-蛋白质相互作用(通过磺基点击化学)34、抗体样蛋白质捕获剂35,36、转录因子37、通道38等的抑制剂也已被表征。
5. 药物输送
药物进入人体的控制给药是有效药物设计的一个重要方面。点击化学已用于构建聚合物纳米和微粒药物递送系统(DDS),如聚合物胶束、脂质体、胶囊、碳纳米管等6,39。
6. 材料优化(聚合物改性)
在材料制造领域,从线性聚合物和接枝聚合物到更复杂结构(如星形聚合物、嵌段共聚物和树状聚合物)的合成,再到表面和界面的功能化40,点击化学都产生了巨大的影响。例如,由于不产生小分子副产物,点击化学可以最大限度地减少气泡、空穴和不规则的形成,就像其他缩聚反应一样,这些气泡、空穴和不规则会破坏新合成热固性材料的外观和性能41。
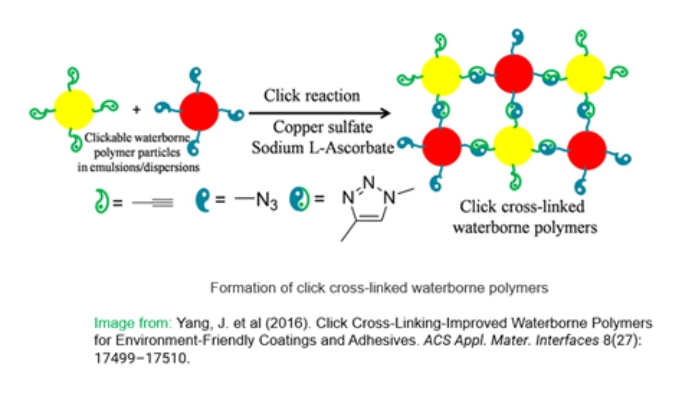
CuAAC click chemistry还被用作一种高效、环保的交联策略,以改善适用于涂料和粘合剂的水性聚合物的性能42(下图2)。广泛适用于聚氨酯(WPU)、聚酯分散体(PED)和聚丙烯酸酯乳液(PAE),该策略优于其他可用的交联策略(包括基于N-羟甲基丙烯酰胺(NMA)、悬垂乙酰乙酸基团和可逆酮酰肼反应的自交联系统)。Click交联聚合物薄膜的机械强度、硬度和耐水/溶剂性能显著提高,为工业涂料应用中使用硬化剂提供了一种有可能降低成本的替代品。
此外,各种(1D、2D、3D)生物材料(如水凝胶)的合成在组织工程43,44,45,46再生医学47、药物输送48和基因治疗领域49也越来越受到重视。
7. 病毒研究探针
在过去几十年中50,与病毒相关的研究,包括病毒(蛋白质、核酸或病毒粒子)追踪51,52、抗病毒药物设计53,54、诊断55,56,57和基于病毒的传递系统58,59都使用了点击化学。例如,通过将叠氮化物修饰的病毒粒子连接到由二苯并环辛烯(DBCO)衍生的量子点(QD),使用无铜点击反应来标记包膜病毒(痘苗病毒(VACV)和禽流感A病毒(H9N2))。标记效率达到80%以上,不干扰病毒的感染能力,荧光强度足以实现单个病毒粒子的跟踪60。
8. CRISPER-sgRNA合成与靶基因标记
Click chemistry现在可以在CRISPR工具箱中找到合成单个或多个单一导向RNA(sgRNA)的位置,绕过了与(更长)寡聚体长度相关的现有合成限制,并缩短sgRNA设计和应用之间的时间。
Click chemistry(被称为“分裂和点击”)不是一次性制造整个sgRNA,而是简单地连接两个更小(更容易合成)的片段:一个按需制备的高纯度~20-mer(crRNA)靶向序列和一个通用的可大规模生产的79-mer CRISPR内切酶蛋白(Cas9)序列(tracrRNA)。结果发现,带有三唑键的~99-聚体能够在体外和细胞内有效地进行Cas9介导的DNA切割,其靶向性与体外转录的sgRNA相当61。
点击化学也被用于标记靶基因(称为sgRNA点击(sgR CLK))62。该技术包括在体外转录的CRISPR-sgRNA的3′端安装点击手柄,以形成叠氮化物标记的三元复合物(由dCas9、sgRNA和靶基因组成)。然后通过与炔烃对应物的点击反应实现该三元络合物的功能化。
此外,点击化学还用于设计一种柔性树枝状聚合物,用于传递锌指、TALEs和CRISPR/dCas9平台。使用该方法具有高转染效率和较大的处理量63。
9. 包括“点击发布”的新应用
除了连接,点击化学现在正在探索解封或“点击释放”应用,这使得探针激活和治疗传递的新策略成为可能64,65,66。例如,利用逆电子需求Diels-Alder-哒嗪消除反应在体外和肿瘤小鼠中激发阿霉素从抗体-药物结合物(ADC)中的快速释放67。
点击化学还被用于开发最先进的微芯片和毛细管系统68,如微流控“点击芯片”69和基于石墨烯的“点击芯片”70。此外,“电点击”接合方法已被用于固定酶(用于生物传感器)、制备电化学免疫传感器以及在空间和时间上控制蛋白质接合71,72,73。
相关阅读
常见点击反应(Click Reactions)及其试剂——CuAAC常见点击反应(Click Reactions)及其试剂——SPAAC
参考文献
- Sharpless et al. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40(11): 2004-2021.
- Liu, B., Kenry. (2019) Bio-orthogonal Click Chemistry for In Vivo Bioimaging. Trends in Chemistry 1(8): 763-778.
- Best, M. D., et al. (2019) Labeling of phosphatidylinositol lipid products in cells through metabolic engineering by using a clickable myo-inositol probe. Chembiochem 20(2): 172-180.
- Li, H., Aneja, R., Chaiken, I. (2013) Click Chemistry in Peptide-Based Drug Design. Molecules 18(8): 9797-9817.
- Prescher, J. A., Dube, D. H., Bertozzi, C. R. (2004) Chemical remodelling of cell surface in living animals. Nature 430: 873–877.
- Agnew B. et al. (2011) Metabolic Labeling and Click Chemistry Detection of Glycoprotein Markers of Mesenchymal Stem Cell Differentiation. In: Vemuri M., Chase L., Rao M. (eds) Mesenchymal Stem Cell Assays and Applications. Methods in Molecular Biology (Methods and Protocols), vol 698. Humana Press.
- Fantoni, N. F., El-Sagheer, A. H., Brown, T. (2021) A Hitchhiker’s Guide to Click Chemistry with Nucleic Acids. Chemical Reviews Article ASAP.
- Das, S. R., Paredes, E. (2010) Click Chemistry for Rapid Labeling and Ligation of RNA. ChemBioChem 12(1): 125-131.
- Lei, X. et al. (2013) A Bioorthogonal Ligation Enabled by Click Cycloaddition of o-Quinolinone Quinone Methide and Vinyl Thioether. J. Am. Chem. Soc. 135(13): 4996-4999.
- Takayama, Y., Kusamori, K., Nishikawa, M. (2019) Click Chemistry as a Tool for Cell Engineering and Drug Delivery. Molecules 24(1):172.
- Salic, A., Mitchison, T. J. (2008) A Chemical Method for Fast and Sensitive Detection of DNA Synthesis in Vivo. Proc. Natl. Acad. Sci. USA 105: 2415-2420.
- Neef, A. B., Luedtke, N. W. (2011) Dynamic Metabolic Labeling of DNA in Vivo with Arabinosyl Nucleosides. Proc. Natl. Acad. Sci. USA 108: 20404-20409.
- Pickens, C. J. et al. (2018) Practical Considerations, Challenges, and Limitations of Bioconjugation via Azide-Alkyne Cycloaddition. Bioconjugate Chem. 29(3): 686-701.
- Bertozzi, C. R. (2008) In vivo imaging of membrane-associated glycans in developing zebrafish. Science 320(5876): 664-667.
- Partridge, A. W. et al. (2021) NanoClick: A High Throughput, Target-Agnostic Peptide Cell Permeability Assay. ACS Chem. Biol. 16(2): 293-309.
- Devaraj, N. K. et al. (2013) Fluorescent live-cell imaging of metabolically incorporated unnatural cyclopropene-mannosamine derivatives. ChemBioChem 14: 205-208.
- Lemke, E. A. (2014) Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed. Engl 53(8): 2245–2249.
- Zhang, X. et al. (2015) Second generation TQ-ligation for cell organelle imaging. ACS Chem. Biol. 10: 1676-1683.
- Chin, J. W. et al. (2014) Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J. Am. Chem. Soc. 136: 7785-7788.
- Xie, H. Y. (2017) Integrating two efficient and specific bioorthogonal ligation reactions with natural metabolic incorporation in one cell for virus dual labeling. Anal. Chem. 89(21): 11620-11627.
- Castro, V., Rodriguez, H., Albericio, F. (2016) CuAAC: An Efficient Click Chemistry Reaction on Solid Phase. ACS Comb. Sci. 18(1): 1-14.
- Gehringer, M., Forster, M., Laufer, S. A. (2015) Solution-Phase Parallel Synthesis of Ruxolitinib-Derived Janus Kinase Inhibitors via Copper-Catalyzed Azide-Alkyne Cycloaddition. ACS Comb. Sci. 17(1): 5-10.
- Chang, Y-T. et al. (2011) Solid phase combinatorial synthesis of a xanthone library using click chemistry and its application to an embryonic stem cell probe. Chem. Commun. 47: 7488-7490.
- Meier, M. A. R. et al. (2019) Direct comparison of solution and solid phase synthesis of sequence-defined macromolecules. Polym. Chem. 10: 3859-3867.
- Zhan, P. et al. (2016) Discovery of bioactive molecules from CuAAC click-chemistry-based combinatorial libraries. Drug Discovery Today 21(1): 118-132.
- Sharpless, K. B. et. al. (2002) Click Chemistry in Situ: Acetylcholinesterase as a Reaction Vessel for the Selective Assembly of a Femtomolar Inhibitor from an Array of Building Blocks. Angew. Chem., Int. Ed. 41, 1053– 1057.
- Oueis, E., Sabot, C., Renard, P.-Y. (2015) New insights into the kinetic target-guided synthesis of protein ligands. Chem. Commun. 51: 12158-12169.
- Hirsch, A. K. H. et al. (2018) Druggability Assessment of Targets Used in Kinetic Target-Guided Synthesis. J. Med. Chem. 61(21): 9395-9409.
- Kolb, H. C. et al. (2004) In Situ Click Chemistry: Enzyme Inhibitors Made to Their Own Specifications. J. Am. Chem. Soc. 126(40): 12809-12818.
- Kolb, H. C. et al. (2005) In Situ Click Chemistry: Enzyme-Generated Inhibitors of Carbon Anhydrase II. Angew. Chem. Int. Ed. 44(1): 116-120.
- Fokin, V. V. et al. (2006) Inhibitors of HIV‐1 Protease by Using In Situ Click Chemistry. Angew. Chem. Int. Ed. 45(9): 1435-1439.
- Hirose T, Sunazuka T, Omura S. (2010) Recent development of two chitinase inhibitors, Argifin and Argadin, produced by soil microorganisms. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences 86(2):85-102.
- Dervan, P. B., Poulin-Kerstien, A. T. (2003) DNA-Templated Dimerization of Hairpin Polyamides. J. Am. Chem. Soc. 125(51): 15811-15821.
- Manetsch, R. (2011) Screening of protein-protein interaction modulators via sulfo-click kinetic target-guided synthesis. ACS Chemical Biology 6(7): 724–732.
- Heath, J. R. (2009) Iterative in situ click chemistry creates antibody-like protein-capture agents. Angew. Chem. Int. Ed. 48(27): 4944-4948.
- Heath, J. R. et al. (2013) In situ click chemistry: from small molecule discovery to synthetic antibodies. Integr. Biol. 5(1): 87-95.
- Deprez, B et al. (2010) Exploring Drug Target Flexibility Using in Situ Click Chemistry: Application to a Mycobacterial Transcriptional Regulator. ACS Chem. Biol. 5: 1007-1013.
- Fokin, V. V. et al. (2012) Generation of Candidate Ligands for Nicotinic Acetylcholine Receptors Via in Situ Click Chemistry with a Soluble Acetylcholine Binding Protein Template. J. Am. Chem. Soc. 134: 6732-6740.
- Fernandez-Megia, E. et al. (2012) Click Chemistry for Drug Delivery Nanosystems. Pharm. Res. 29: 1-34.
- List-Kratochvil, E. J. W. et al. (2020) Utilizing Diels-Alder “click” chemistry to functionalize the organic-organic interface of semiconducting polymers. J. Mater. Chem. C. 8: 3302.
- Serra, À. et al. (2020) The Use of Click-Type Reactions in the Preparation of Thermosets. Polymers 12(5):1084.
- Yang, J. et al. (2016) Click Cross-Linking-Improved Waterborne Polymers for Environment-Friendly Coatings and Adhesives. ACS Appl. Mater. Interfaces 8(27): 17499-17510.
- DeForest, C. A., Anseth, K. S. (2011) Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat. Chem. 3(12): 925-931.
- Becker, M. L. et al. (2013) Postelectrospinning “click” modification of degradable amino acid-based poly (ester urea) nanofibers. Macromolecules, 46(24): 9515-9525
- Chen, H.-Y. et al. (2016) Multifaceted and route-controlled “click” reactions based on vapor-deposited coatings. Biomater. Sci. 4(2): 265-271.
- Xu, Z., Bratlie, K. M. (2018) Click Chemistry and Material Selection for in Situ Fabrication of Hydrogels in Tissue Engineering Applications. ACS Biomater. Sci. Eng. 4(7): 2276-2291.
- Shi, L. et al. (2021) Click chemistry-based biopolymeric hydrogels for regenerative medicine. Biomedical materials 16(2): 022003.
- Fu, Q. et al. (2017) Clickable and imageable multiblock polymer micelles with magnetically guided and PEG-switched targeting and release property for precise tumor theranosis. Biomaterials 145: 138-153.
- Lin, C. et al. (2020) Bioreducible crosslinked cationic nanopolyplexes from clickable polyethylenimines enabling robust cancer gene therapy. Nanomedicine: Nanotechnology, Biology and Medicine 24: 102144.
- Ren, L. et al. (2018) Recent trends in click chemistry as a promising technology for virus-related research. Virus research 256: 21–28.
- de Haan, C. A. et al. (2012) Visualizing coronavirus RNA synthesis in time by using click chemistry. Journal of virology 86(10): 5808–5816.
- Kalveram B, Lihoradova O, Indran SV, Head JA, Ikegami T. (2013) Using click chemistry to measure the effect of viral infection on host-cell RNA synthesis. Journal of Visualized Experiments: Jove.
- Miura Y. et al. (2017) Design of glycopolymers carrying sialyl oligosaccharides for controlling the interaction with the influenza virus. Biomacromolecules 18(12):4385–4392.
- Wang, C., Zhu, W., Wang, B.Z. (2017) Dual-linker gold nanoparticles as adjuvanting carriers for multivalent display of recombinant influenza hemagglutinin trimers and flagellin improve the immunological responses in vivo and in vitro. Int. J. Nanomed. 12: 4747-4762.
- Donolato, M. et al. (2015) Quantification of NS1 dengue biomarker in serum via optomagnetic nanocluster detection. Scientific Reports 5: 16145.
- Samitier J. et al. (2015) Label-free electrochemical DNA sensor using "click"-functionalized PEDOT electrodes. Biosens. Bioelectron. 74:751-756.
- Carell, T. et al. (2020) Supersensitive Multifluorophore RNA-FISH for Early Virus Detection and Flow-FISH by Using Click Chemistry. ChemBioChem 21(15): 2214-2218.
- Iyer, K. S. et al. (2017) Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chemical science 8(4): 2923–2930.
- Chu, Y., Oum, Y. H., Carrico, I. S. (2016) Surface modification via strain-promoted click reaction facilitates targeted lentiviral transduction. Virology 487: 95–103.
- Xie, H. et al. (2012) A Mild and Reliable Method to Label Enveloped Virus with Quantum Dots by Copper-Free Click Chemistry. Analytical chemistry 84: 8364-8370.
- Brown, T. et al. (2019) An artificial triazole backbone linkage provides a split-and-click strategy to bioactive chemically modified CRISPR sgRNA. Nat Commun 10: 1610.
- Srivatsan, S. G. et al. (2020) Terminal Uridylyl Transferase Mediated Site-Directed Access to Clickable Chromatin Employing CRISPR-dCas9. J. Am. Chem. Soc. 142(32): 13954-13965.
- Iyer, K. S. et al. (2017) Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chemical science 8(4): 2923–2930.
- Carlson, J. C. T., Mikula, H., Weissleder, R. (2018) Unraveling Tetrazine-Triggered Bioorthogonal Elimination Enables Chemical Tools for Ultrafast Release and Universal Cleavage. J. Am. Chem. Soc. 140(10): 3603-3612.
- Royzen, M. et al. (2016) In Vivo Bioorthogonal Chemistry Enables Local Hydrogel and Systemic Pro-Drug To Treat Soft Tissue Sarcoma. ACS Cent. Sci. 2(7): 476-482.
- Peplow, M. (2019) Click chemistry targets antibody-drug conjugates for the clinic. Nature Biotechnology 37: 835-837.
- Robillard, M. S. et al. (2016) Triggered Drug Release from an Antibody-Drug Conjugate Using Fast “Click-to-Release” Chemistry in Mice. Bioconjugate Chem. 27(7): 1697-1706.
- Chen, C. et al. (2019) Click chemistry at the microscale. Analyst 144: 1492-1512.
- Reichert, D. E. et al. (2015) Development of a microfluidic “click chip” incorporating an immobilized Cu(I) catalyst. RSC Adv. 5: 6142-6150.
- Aran, K. et al. (2018) Graphene-based biosensor for on-chip detection of bio-orthogonally labeled proteins to identify the circulating biomarkers of aging during heterochronic parabiosis. Lab Chip 18: 3230-3238.
- Pingarrón, J. M. et al. (2020) Design of electrochemical immunosensors using electro-click chemistry. Application to the detection of IL-1β cytokine in saliva. Bioelectrochemistry 133: 107484.
- Ono, T. et al. (2019) Enzyme immobilization in completely packaged freestanding SU-8 microfluidic channel by electro click chemistry for compact thermal biosensor. Process Biochemistry 79: 57-64.
- Shi, X-W. et al. (2013) Protein addressing on patterned microchip by coupling chitosan electrodeposition and ‘electro-click’ chemistry. Biofabrication 5(4): 041001.